

Physik Department E22
Technische Universität München
James-Franck-Str. 1
85748 Garching
Germany
Room 3111
+49 89 289 124 95
Physik Department E22
Technische Universität München
James-Franck-Str. 1
85748 Garching
Germany
Room 3111
+49 89 289 124 95
Thomas Suren
Thomas Suren

Molecular Machines
Proteins are fundamentally important to life, since they perform all of functions within living organisms. It is the protein’s structure that defines its biological function within a cell, however, protein function is a dynamical process. Therefore, to understand completely how a protein functions, we must also understand how it moves.
Using single molecule measurements of proteins, it is possible to directly observe and manipulate protein motions, and to study how they are influenced by interactions with other molecules. From such experiments, the rates of folding, unfolding, association and dissociation as well as stabilities of the proteins under different conditions can also be found.
The experimental systems used in the lab, custom-built dual-beam optical traps, provide sub-nanometre spatial and pico-Newton force resolution. Using this setup, in combination with biochemical functional assays, molecular machines are studied in detail.
Molecular chaperones are essential proteins for cell viability, which assist in the folding, unfolding, assembly and disassembly of their client proteins. They can also maintain protein stability under conditions of stress. Several different chaperones are studied in the Rief group.
Heat shock protein 90 (Hsp90) is a molecular chaperone which is one of the most abundant proteins found in eukaryotic cells. Its abundance underlines its importance, as it is known to be involved in the assembly and regulation of cellular signalling systems which, in turn, ensure that cell growth is regulated.
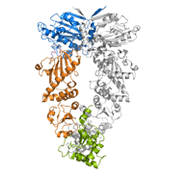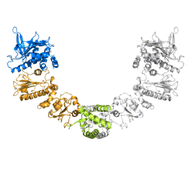
The open and closed conformations of Hsp90 from crystal structures (PDB accession codes: 2CG9 and 2IOQ)
For measuring force one needs two things: a spring and calibration. Both are provided by our “dual beam optical tweezers setup”.
Due to momentum conservation it is possible to trap microscopic particles using sharply focused laser beams. The object trapped in the laser focus obeys Hooke’s law, where the restoring force felt by the object is linear to ist displacement from the trap centre. Thus, the focused laser beam can be seen as a spring for microscopic objects.
We use this effect to trap two 1µm-diameter silica beads, functionalized in such a way that we can hold a single protein molecule between them. By moving one of the laser beams by very small, accurately-controlled nm-scale distances, we can apply piconewton (pN) forces to individual protein molecules.

E.coli Hsp70: DnaK structure from NMR (PDB accession code: 2KHO)